Intensive care is an ideal environment for the use of Big Data Analysis (BDA) and Machine Learning (ML), due to the huge amount of information processed and stored in electronic format in relation to such care. These tools can improve our clinical research capabilities and clinical decision making in the future.
The present study reviews the foundations of BDA and ML, and explores possible applications in our field from a clinical viewpoint. We also suggest potential strategies to optimize these new technologies and describe a new kind of hybrid healthcare-data science professional with a linking role between clinicians and data.
La gran cantidad de información que se procesa informáticamente en el entorno de la medicina intensiva la convierte en un campo ideal para el empleo de técnicas conocidas como Big Data Analysis (BDA) y Machine Learning (ML), que pueden permitir en el futuro mejorar nuestra capacidad de investigación clínica y dirigir de manera más precisa las terapias que proporcionamos a nuestros pacientes.
En este artículo se revisan los conceptos fundamentales sobre BDA y ML, y se estudian sus posibles aplicaciones al ámbito de la medicina intensiva, desde un punto de vista del clínico. También se plantean potenciales estrategias para sacar el máximo partido a estas tecnologías emergentes, incluyendo la aparición de un nuevo tipo de profesional sanitario encargado de actuar como enlace entre la parte clínica y la ingeniería de datos.
As professionals in intensive care medicine, we live immersed in a sea of data. In a digitalized Department of Intensive Care Medicine such as that of Hospital Clínico San Carlos (Madrid, Spain) (with three Intensive Care Units [ICUs] and approximately 2400 admissions a year), an average of 1400 new units of information are entered in the electronic database per admitted patient on a normal working day – this implying about 10 million units of information a year (Núñez A., personal communication).
The exponential development of computing technology and the incorporation of systems with great storage and processing capacities that can be acquired at an accessible cost imply the registry of a great volume of information that can be used in different ways. Even those Units that still lack electronic case histories or intensive care software applications can take advantage of the latest-generation computing methods to improve the way in which daily work is done. An example is the application of Natural Language Processing (NLP) to a series of reports filed in MS Word, PDF or other similar free-text (non-structured) formats.1
The data managed by intensivists can come from different sources. Health professionals continuously add information (whether structured or not) to the documentation of the patient. It has been estimated that physicians spend almost 2h documenting for every hour of direct patient care.2 Physicians, nurses and technicians do this in the form of free-text notes or reports of different specialties, or as coded data referred to diagnoses or procedures. We also generate treatment instructions and drug administration registries, and receive a large body of data produced by medical devices and systems: laboratory analyzer results, vital signs, advanced monitoring data, ventilator parameters, operating parameters of complex equipment such as perfusion pumps, dialysis monitors, extracorporeal membrane oxygenation (ECMO) systems, and information in the form of images, audio, video and much more. Until only a few years ago, all this information was lost or, at most, filed as case histories in paper format. Nowadays, however, it is possible to store and process this information automatically in digital format and use it to extract new knowledge and obtain guidance for improved patient care.
There is a broad range in the degree to which the data we use are structured. It is much easier to conduct clinical research when the information is available in a structured format, though health professionals have not yet reached consensus regarding an unequivocal way to express each health-related concept. This does not mean that we do not have standards. The SNOMED CT,3 HL7,4 UMLS,5 DICOM,6 LOINC7 and many others are examples of such standards, and are used in different healthcare settings – allowing automated processing of data and the exchange of information between systems. But standards are not such in absolute terms: we can express a given diagnosis using different standards, such as MESH,8 UMLS, SNOMED CT, ICD-99 or ICD-10,9 and it often proves necessary to establish “translations” between standards (the technical term for this being “mapping”) in order to transfer information from one system to another. Solutions involving new approaches to the problem of multiple standards (for example OMOP10) offer us tools for performing such mapping processes in a systematic way.
Not only data are important. The context in which the information is set is also important. In some cases a piece of information has no value at all unless we can associate it to more relevant information about the patient or the clinical situation. As an example, a blood pressure recording of 90/60mmHg is interpreted very differently in a young woman undergoing plastic surgery versus a hypertensive elderly patient with bleeding. One same concept moreover can be reflected in different ways by different professionals, in different situations and – in the case of numerical values – using different units.
Our structured data can be categorical, consist of whole numbers or with decimals, comprise dates, hours or durations, or may be grouped (e.g., blood pressure with its two components: systolic and diastolic) in a way similar to the lists, tuples or objects we see as data structures in programming languages. Sometimes our data are matrixes of bytes organized to represent an image or video, such as for example a DICOM file that can be used to display a computed tomography (CT) image. In this concrete example there is another interesting concept, namely “metadata”: information about the information (e.g., in the case of the CT scan, the image acquisition parameters would represent metadata).
Some data are subjective. As an example, a physician may reason the way in which he or she reaches a certain diagnosis through phrases in a free-text note. In contrast, other data are objective: our hemofilter has extracted exactly 52ml of ultrafiltrate from the patient during the last hour, and this information is conveyed to the electronic plot via the network connections.
In performing secondary analyses of clinical data we must consider a number of aspects referred to privacy, security and sensitivity. It may be perfectly acceptable for a patent to know that his or her respiratory frequency is publicly disclosed, but the same possibly cannot be said of seropositivity status for the human immunodeficiency virus.
Data are of different origins because they are obtained for different reasons (accountancy, research or clinical management of the patient, for example), and so there are also different actors involved in the process (managers, clinicians, researchers, service suppliers).
If we wish to extract valid information from all this, with a view to helping health professionals to make better decisions and guide our clinical research, we must be able to work efficiently with this clinical jigsaw puzzle of thousands of small pieces generated by daily patient care.
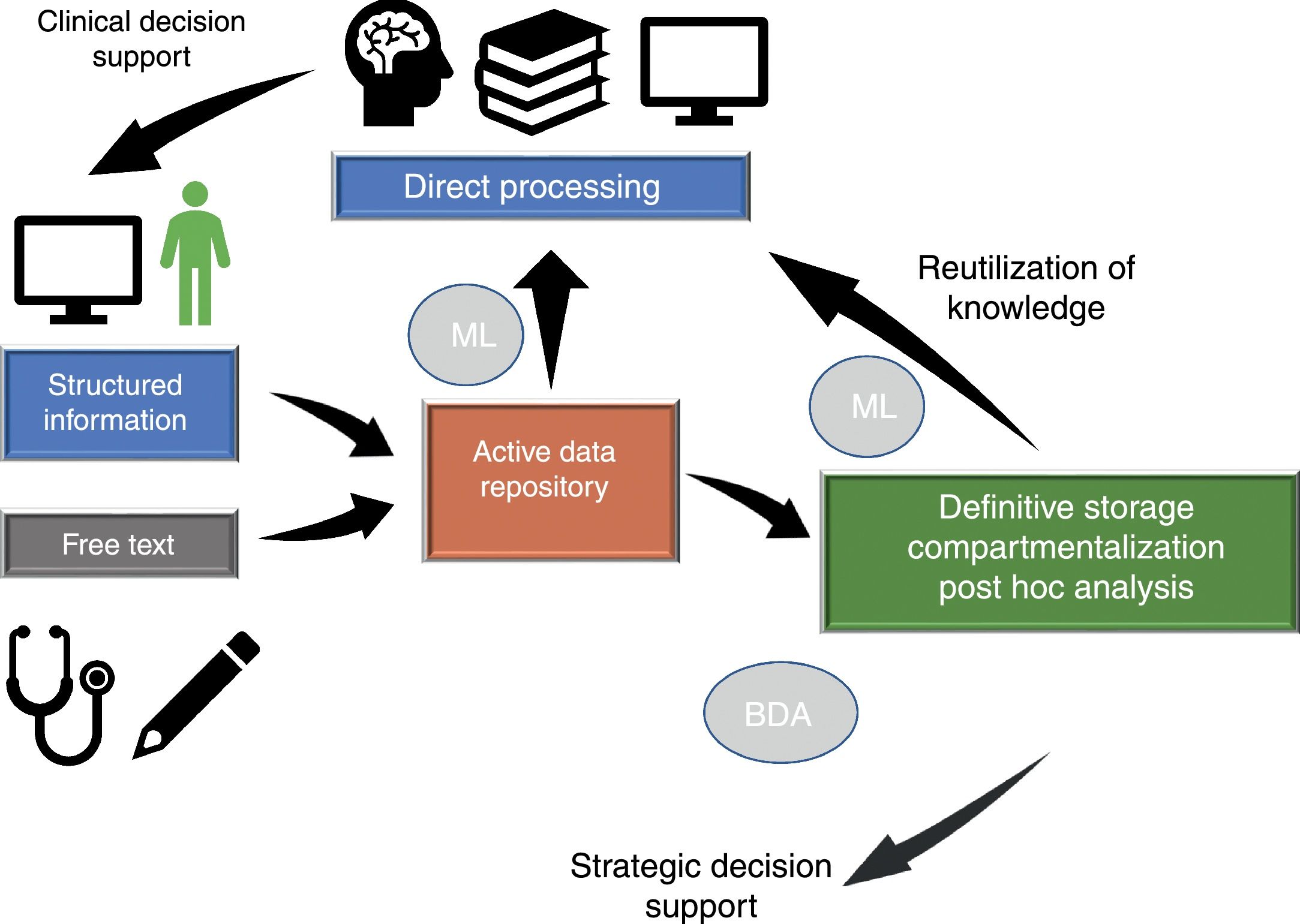
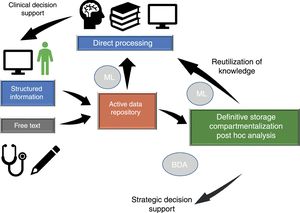
Dataflow in intensive care medicineInformation flows in our setting through different phases which we will briefly consider (Fig. 1), since in different moments of the clinical care process critical events occur that influence our capacity to make efficient use of the information.
Dataflow in intensive care medicine. The quality of care and of strategic decision making depends on processing of the information contained in the active data repository (information that is being generated in each moment and which is available for decision making) and on reutilization of the knowledge previously stored in local or shared databases. The automated processing of information through Machine Learning (ML), and the possibility of rapid access to large volumes of data of a heterogeneous structure by means of Big Data Analysis (BDA), allow us to improve the care processes and extract knowledge from the data.
A part of the data input process is performed automatically and in a structured format allowing for efficient information use. However, there is a great opportunity for improvement in the acquisition of data in which free text is currently used. We will try to explain this with an example: “A 58-year-old male reporting one hour ago to the emergency room due to dyspnea and central chest pain irradiating to the left arm…”.
In this brief paragraph of 23 words there are at least 8 concepts that can be expressed in a structured manner in the following xml document (a standard format for displaying information11):
=“years”>58
For a physician it is usually much easier to interpret the free text paragraph, while in the case of a computer system the second way of representing the information is much more efficient. The ideal situation would be to develop a tool allowing the human to enter the information at least as fast and conveniently as with the free text, but also allowing direct input in a structured format, facilitating reading and understanding of the data by both humans and computers.
Processing of the data in vivoThe data entered in the system is in a “passive” state, and it is the health professional who must make the pertinent inferences after compiling the data that he or she feels to be relevant in a concrete moment of the patient course.
However, we have the possibility of transforming the filed information into “active” data capable of triggering system responses through a so-called Clinical Decision Support System (CDSS). Continuing with the previous example, suppose we have entered the above paragraph, and the system has a concept on file:
By entering the patient information, the system may alert us directly to the fact that the patient has a symptom consistent with acute coronary syndrome. This is a very simple example, but what is really useful about the software tools is that this type of processing can be done automatically for thousands of concepts and in a much more systematic way than is done by the human brain. To understand this by means of an analogy, it is as if the system were running a continuous checklist with our patients. In addition, the current artificial intelligence (AI) techniques allow the use of experience gained with previous patients in the form of structured data for the assessment of the next individual patient. In future, this may have a great impact upon clinical practice.
Another example is the use of real time control systems, for example to continuously adjust the insulin perfusion dose of a patient according to the insulin sensitivity he or she has shown previously; stress condition assessed from different data of the plots and laboratory tests; and the caloric and carbohydrate supply being provided in that moment. Over time, the system can learn about the patient and gradually optimize glycemic control within concrete safety parameters.
Data storageThere are two ways to store the information of our patients: (a) in relational databases that use the SQL (Structured Query Language) to retrieve and process the stored information; and (b) in non-structured data repositories (NoSQL). Nowadays there are tools that allow the extraction and processing of information also from data of this kind. Specific types of information can be stored in specialized databases such as for example a PACS for the filing of clinical images, using the DICOM format.
A posteriori data analysis and the sharing of informationOnce we have stored the data, they can be processed in different ways. For example, we can obtain an automatic report on the activity of our Unit by consulting our SQL database with the demographic and clinical information of our patients, or we can find out which antibiotics we use most often, or what type of disease conditions are predominantly seen in our Unit, without having to again review our patients one by one. This information can be used for benchmarking (comparison of results between Units or within one same Unit when processes or resources are modified), or for planning strategic actions.
We recommend the article published by Celi et al.,12 where a review is made of all this dataflow cycle and the concept of “closing the loop” is addressed, explaining the way in which AI can contribute to the development of intensive care medicine.
What is Big Data?The classical approach of the clinician to the management of data for clinical research is based on the premise that it is necessary to obtain quality information in order to secure reliable results that are applicable to patients. The difficulty of obtaining such information leads to attempts to optimize the process, applying prospective designs, randomization and a working hypothesis before the data are analyzed.
As an alternative to the exclusive use of data collected in an orthodox manner, Big Data Analysis (BDA) offers the novelty of detecting the underlying structure and knowledge in large bodies of information, even when the latter do not seem to be structured.
A popular definition of the concept13 is that “Big Data consists of data sets with such a large volume and such a broad structural variety that specific technology and analytical methods are needed to process them and transform them into knowledge or value”.
Specific BDA techniques have been successfully used in fields such as marketing, strategic decision making in the business world, banking, transport, logistics, insurance or the detection of fraud in electronic commerce. There are no reasons to believe that BDA cannot be applied to our setting, where we continuously make strategic or concrete decisions in given patients whose characteristics – while complex – are often repetitive.
A recent example, which we will address again further below, is the follow-up of influenza epidemics based on analysis of the searches made on the internet. In this case we use databases with many millions of registries in order to draw rapid and reliable epidemiological conclusions.14
The open code philosophy has acquired a strong presence in the world of Big Data, allowing its use without the need for major economical investments. Projects such as the Apache Hadoop15,16 include a whole range of resources in their ecosystem (HDFS, Spark, MapReduce, Impala, HBase, Hive) that allow the low-cost construction of a Big Data batch processing system (analysis of data already stored in large relational or non-relational databases) or Big Data stream processing system (analysis and processing of data as they are generated).
What is Machine Learning?The concept of Machine Learning (ML) dates back to the mid-twentieth century, and was defined in an article published by Samuel in 195917 as an area of AI that uses computational algorithms and statistics to give computers the capacity to “learn”, i.e., to improve their results in a specific task after processing a sufficient volume of information and without explicit external instructions (which may be biased) from the programmer.
The scope of ML is closely related to other fields such as simulation and modeling, the optimization of systems and statistics. All of these areas make intensive use of common mathematical techniques that require specific training.
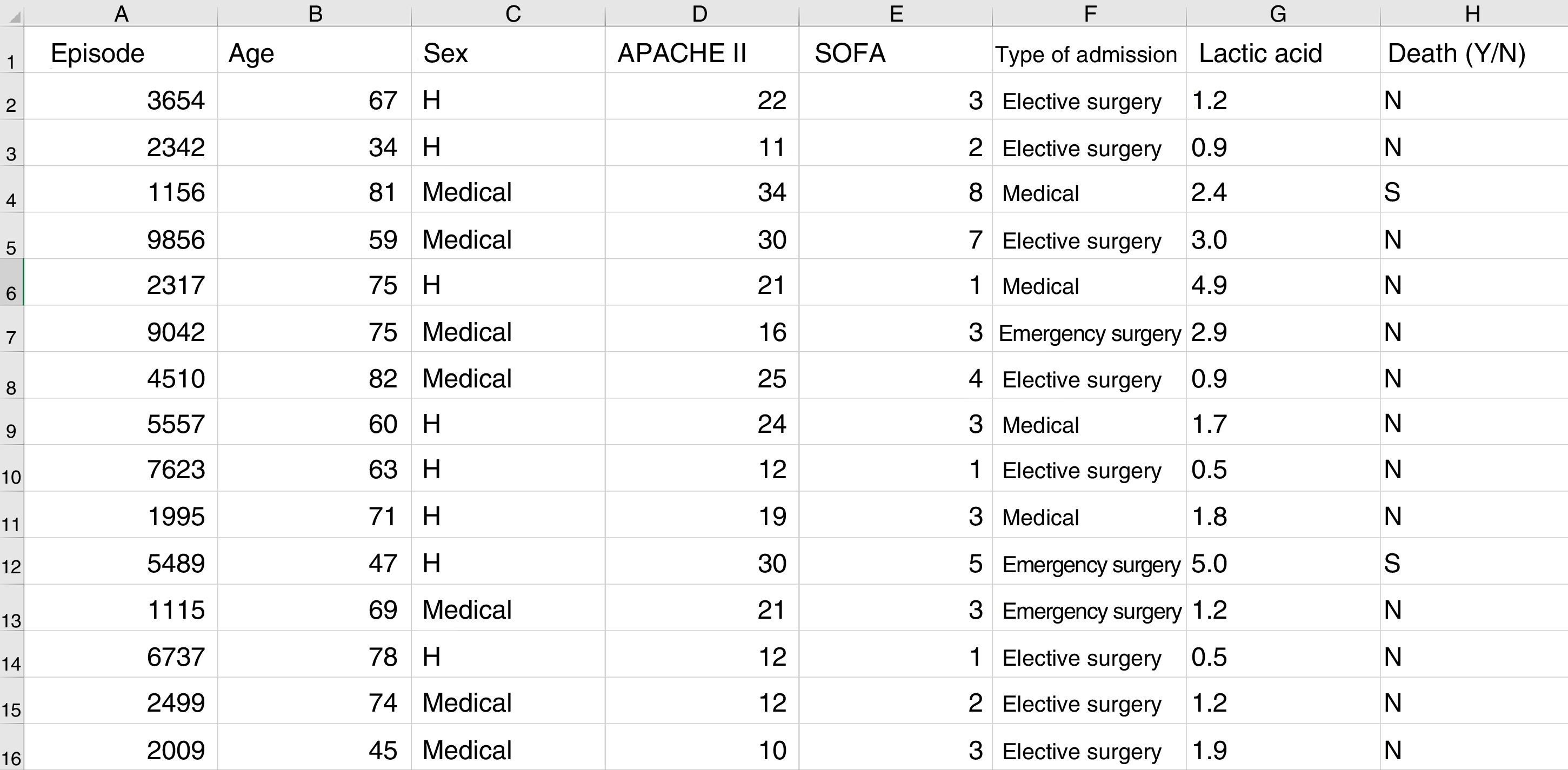
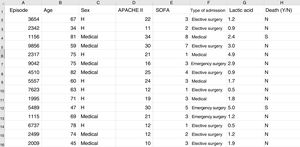
Some basic concepts need to be explained at this point. We must use a format referred to as a “data frame” to work with data in ML. A data frame is a matrix where each row corresponds to one of the patients or registries, and each column corresponds to one of the recorded variables (Fig. 2).
Typical structure of a data frame, composed of rows for each of the registries, with an anonymized identifier, and columns corresponding to the values of different variables for each case. The last column shows the target variable (in this case mortality). Here we can see a section of some registries of the database used as an example in the material included in the GitHub repository.
As an example, suppose we wish to develop a model for the prediction of mortality according to the following variables: type of admission (medical, elective surgery or emergency surgery), age, sex, APACHE II score upon admission, SOFA score upon admission, and blood lactic acid levels. In this case we have as many rows as there are recorded patients, and each row has a column identifying the episode (on an anonymous basis) and a column for each of the explanatory variables that we have defined, plus another column corresponding to the target variable (in this case mortality as a dichotomic variable [YES/NO]).
For those readers who wish to reproduce the examples provided in this review, we have prepared a series of anonymized data from our own database, and have entered them in an MS Excel table in a GitHub repository (https://github.com/anunezr/revision_medicina_intensiva).
There are three main variants of machine learning:
- •
Supervised learning. Each registry is labeled with a value of the target variable, and use is made of different techniques capable of predicting the value of that variable in a new registry. In our above example, the target variable is mortality (YES/NO). Once the system has been “trained”, it must be able to predict the value of that variable corresponding to an episode that has not been previously presented to the system. Therefore, the training data and the data of the test must be different and stored separately. This variant is used in classification, prediction and similarity detection tasks.
- •
Unsupervised learning. In this case the aim is to detect patterns or trends in data without using a target variable. This variant is used for example to automatically classify patients into groups, and to reduce the number of variables and the complexity of the models.
- •
Reinforcement learning. In this case the system pursues an objective or reward, and progressively learns as the environment with which it interacts is explored, and which is not known beforehand – avoiding actions with a negative reward and seeking to conduct actions with a positive reward. An example of this would be a system that learns and adjusts the empirical antibiotic regimens prescribed by the clinicians as they receive the results and characteristics of the septic patients they see.
Table 1_Sup, included in Annex A of the Supplementary Material, summarizes the different ML techniques for each of these variants. The use of such techniques requires knowledge in programming and control of the concepts used in AI, which normally are not within the reach of the clinician. Because of this, a new collaborative working approach is being consolidated, including the introduction in the Units of a new professional with basic clinical knowledge and advanced skills in statistics and the tools and methods of BDA and ML. These professionals, working jointly with the clinicians, can help us to draw value from a large amount of clinical information which right now is filed in our databases. This “man in the middle”, now already found in leading technological development centers, will soon become an important element in the teams of our Units, and will help to plan strategic decisions and optimize the software tools in order to get the most out of them.
For those readers with knowledge of R or Python and who wish to acquire basic practice in the field of ML, the GitHub repository associated to this review (https://github.com/anunezr/revision_medicina_intensiva) includes a series of scripts that use the different techniques reflected in Table 1_Sup, fundamented on the anonymized information of the database provided as an example.
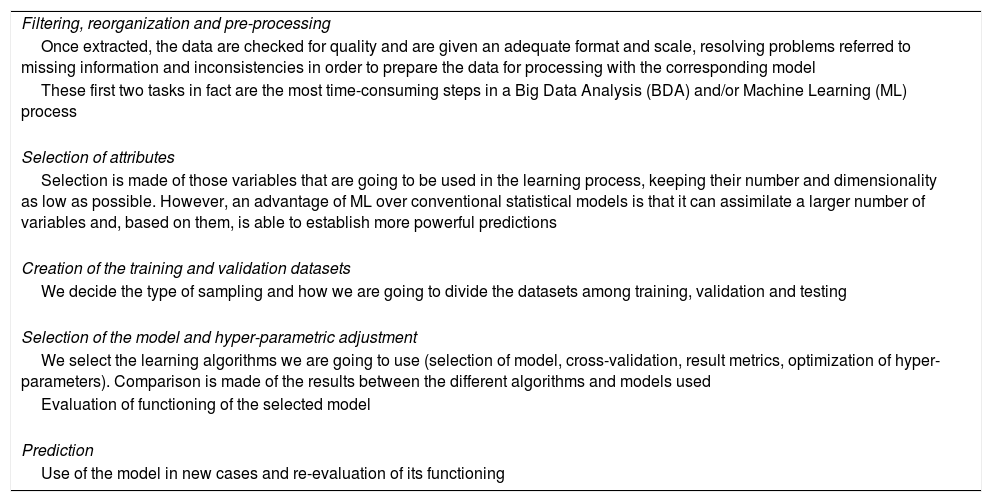
Table 1 provides a “road map” for putting an ML system into practice, adapted to a concrete problem.18 Each of the considered aspects would merit a review as extensive as this article, if not more so. The aim of the table is to explain that the use of BDA and ML requires complex methodology and systematization that in turn demand specific knowledge and experience in order to obtain results. Only under these premises can we extract knowledge from the data in an efficient manner.
Steps in constructing a Machine Learning system.
| Filtering, reorganization and pre-processing |
| Once extracted, the data are checked for quality and are given an adequate format and scale, resolving problems referred to missing information and inconsistencies in order to prepare the data for processing with the corresponding model |
| These first two tasks in fact are the most time-consuming steps in a Big Data Analysis (BDA) and/or Machine Learning (ML) process |
| Selection of attributes |
| Selection is made of those variables that are going to be used in the learning process, keeping their number and dimensionality as low as possible. However, an advantage of ML over conventional statistical models is that it can assimilate a larger number of variables and, based on them, is able to establish more powerful predictions |
| Creation of the training and validation datasets |
| We decide the type of sampling and how we are going to divide the datasets among training, validation and testing |
| Selection of the model and hyper-parametric adjustment |
| We select the learning algorithms we are going to use (selection of model, cross-validation, result metrics, optimization of hyper-parameters). Comparison is made of the results between the different algorithms and models used |
| Evaluation of functioning of the selected model |
| Prediction |
| Use of the model in new cases and re-evaluation of its functioning |
In order to make use of BDA and ML techniques, we must master at least some of the analytical statistical computation languages (R, Python or Java); control SQL as a database consulting tool; and know how to use the code libraries employed in this field. The Supplementary Material provides a brief description of the most widely used software.
Difficulties and strategies for applying these techniques in a Department of Intensive Care MedicineIn the face of this mix of acronyms and technicisms, intensivists tend to ask themselves whether BDA and ML techniques can really be applied in their working environment, and how to do so in an accessible way. In truth, the purpose of this review is to make the clinician aware that entering the world of AI applied to intensive care medicine is possible, but demands structural changes and investment in human resources and technology, as well as an expanded vision of the concept of clinical research.
Logically, the first requirement is a data acquisition and storage system allowing subsequent analytical processing. In Spain we are still far from achieving this, since the existing electronic clinical information systems are not universally implemented in our Units, and an inter-hospital standard is even less common. Furthermore, many of those centers that do have such applications lack efficient access to the stored data – often because the system purchasing contract did not initially contemplate this aspect, which usually comes at an added cost. These paradoxical situations, which border on ethical neglect by keeping clinical data “hostage” of the suppliers of the commercial software, must be urgently resolved if we want to work with the new BDA and ML tools for the good of our patients.
A second consideration is the need for hospitals to assign specialized staff from the Information Technology Department to the custody, processing and analysis of the clinical data of our Units, working in collaboration with the physicians and nurses in order to get the most out of the stored information.
A third consideration is the need to work with the local and regional Ethics Committees to guarantee data security and privacy, and to not obstruct opportunities for improvement of the processes and clinical care which exploitation of the stored data can afford.
Shared databasesSecondary analysis of electronic health records (EHRs)19 refers to processing of the clinical data of patients generated as a subproduct of their acquisition with healthcare purposes, and can be used as an aid in strategic decision making in a Unit, or on a point basis for a concrete patient.
In seeking to endorse clinical research in the intensive care setting, some institutions with very large clinical databases have placed them at the disposal of investigators throughout the world, following due anonymization to preserve data privacy. The currently most popular initiative of this kind is the Medical Information Mart in Intensive Care (MIMIC-III),20,21 the main characteristics of which are its public accessibility and the excellent quality of the filed data, comprising clinical information of all kinds, drawn from over 58,000 critical patients – both adults and newborn infants.
Another example of such databases is the eICU Collaborative Research Database,22 comprising information from a combination of many Intensive Care Units throughout the United States. The registries of the collaborative database compile data from almost 200,000 patients admitted to critical care in 2014 and 2015.
As we can imagine, although such secondary analysis may be very useful for a Department of Intensive Care Medicine or for a clinical investigator, its potential is boosted if the data from several Units can be combined to produce a large multicenter database. The effect is similar to what we see on comparing the conclusions that can be drawn from a single center trial versus a multicenter trial.
However, a number of difficulties must be overcome in order to do this.
Each Unit compiles the data using software that might not be the same as that used in the rest of the Units we seek to merge. We therefore must ensure conversion to a common data structure allowing combined or pooled analysis. This is what we referred to above as “mapping”. In this regard, ontologisms have been introduced that allow us to express and map the filed concepts in a uniform manner. Some of them, such as the Observational Medical Outcomes Partnership (OMOP)10 or Informatics for Integrating Biology and the Bedside (i2b2),23 offer tools to facilitate local database migration to a common data standard.
On only considering the studies published in recent years using the public MIMIC20 database and software libraries with latest-generation algorithms and free access, we can see that interesting results have been obtained in very diverse areas of intensive care medicine (Table 2).
Aspects of clinical research studied using Big Data Analysis and Machine Learning techniques.
| Usefulness of the use of echocardiography in sepsis35 |
| Usefulness of knowing the variability of blood glucose levels in the prognosis of non-diabetic patients36 |
| Improvement in the prediction of in-ICU mortality according to physiological parameters37 |
| Early detection of sepsis38 and the possibility of generating early alerts in patients at a high risk of developing septic shock39 |
| Difference between the water balance targets to be established between the first and the second 24h of sepsis resucitation40 |
| Predictive value of the nursing notes in relation to long term patient prognosis41 |
| Usefulness of early biliary tract drainage in acute cholangitis42 |
| Capacity of natural language processing tools to detect diagnoses not coded in free text notes29 |
| Negative prognostic value of prolonged tachycardia43 |
| Paradoxical relationship between obesity and mortality in the ICU among patients with chronological disease conditions44 |
| Prediction of undesired early readmissions45 |
| Use of additional clinical variables to better adjust heparin perfusion doses46 |
| Poorer prognosis after admission to the ICU among patients administered serotonin reuptake inhibitors47 |
Furthermore, an advantage of this strategy is that the results are fully reproducible: we can take the data set used by the authors and reproduce the statistical and ML methods they have used, even with our own information, since they are routinely available in public repositories such as GitHub.24,25 This causes the studies to be subject to additional quality control, and allows anyone to check both the methods and the results obtained.
There are conditioning elements of a legal and ethical nature that can complicate the data sharing process, and it may prove necessary for the local Ethics Committees to authorize access and check the safekeeping of data security and privacy.
The logistics of the process require coordination among the participating centers and technological and hardware support concordant with the project, involving multidisciplinary teams capable of dealing with the challenges which the different aspects of such a project require.
Once these difficulties have been overcome, the opportunities for improved management, benchmarking, and of progress in clinical research are enormous, and without doubt will compensate the efforts required to incorporate a Department of Intensive Care Medicine to projects of this kind.
Natural language processingSince most of the information which professionals enter in the case history is in the form of free text (also known as natural language), a possible application of AI techniques in medicine is the automated creation of structured information from free text, as well as the classification or phenotyping of patients into different groups according to the contents of the text written by the professionals. These techniques, known as natural language processing (NLP), are advancing quickly.26 There are two possible approaches:
- •
The extraction of concepts from the text using tools such as cTAKES27 is quite well developed in the Anglo-Saxon world, though there are few practical options for texts not written in English. The research group in Data Mining of the Biomedical Technology Center of the Polytechnic University of Madrid (Spain) is working with a tool derived from the cTAKES known as TIDA,28 and in future it is possible that systems of this kind will become part of our computer-based resources.
- •
Another alternative is the use of convolutional neural networks (CNNs),29 but this type of strategy poses an inconvenience in that the algorithms obtained are not easily interpretable by physicians – though this could become irrelevant if the model functions correctly – and the result obtained are variable and conditioned to the sought phenotypes.
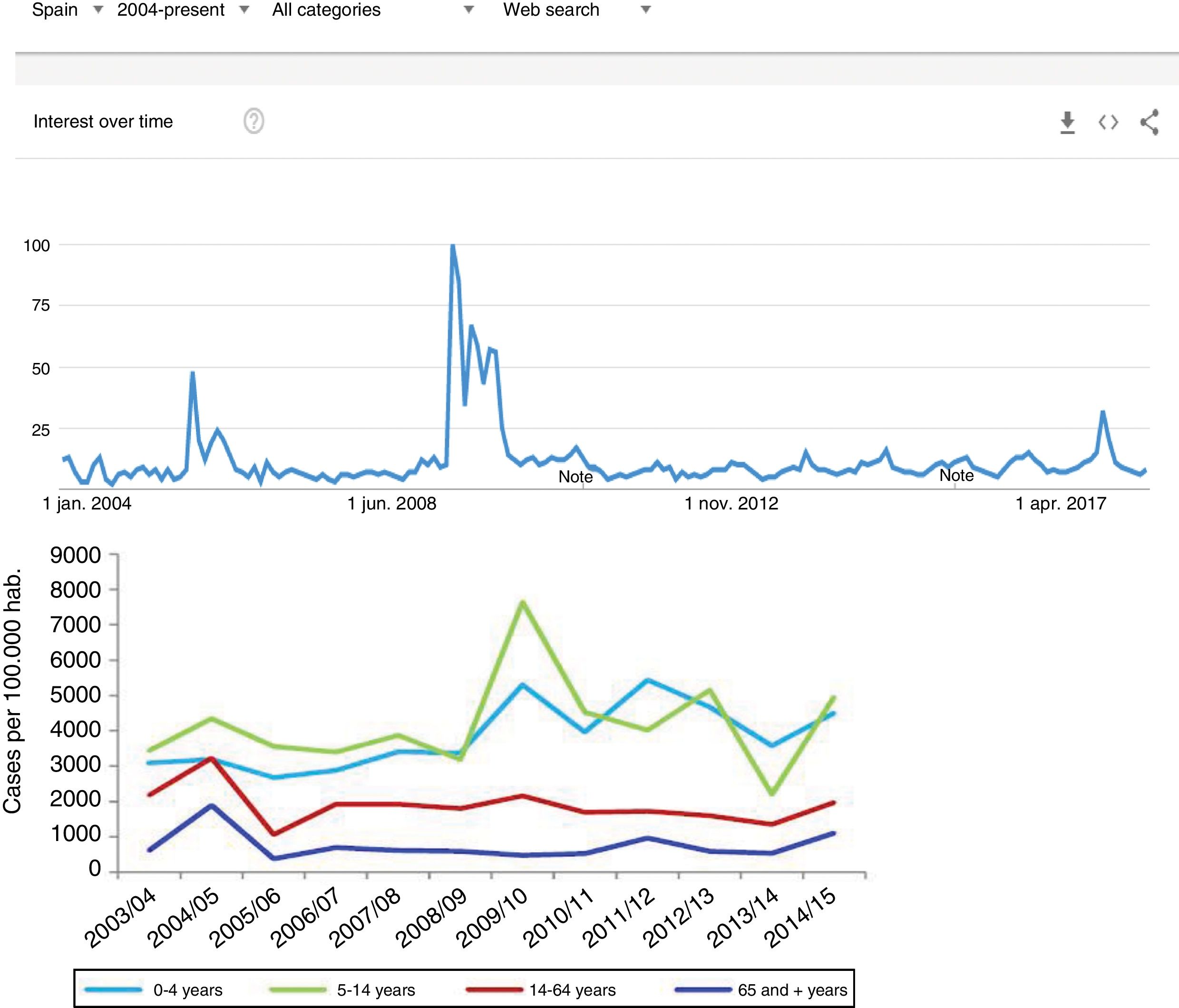
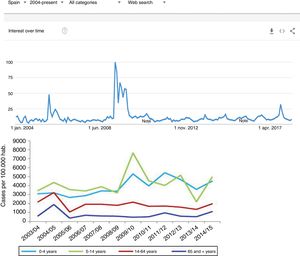
Since the clinical data stored in our clinical information systems are not compiled with the same demanding criteria as in the context of clinical trials, how can the secondary analysis of patient care information allow reliable conclusions to be drawn? They key lies in the volume of the data and in the capacity of the ML tools to detect structure in the midst of caos.19 As an example, we can use follow-up of the incidence of the H1N1 influenza epidemic of 2008 based on the counting of Google searches made using the term “flu”,14 and which has continued to be validated posteriorly (Fig. 3) – exhibiting perfect correlation with the registry of cases obtained by means of more orthodox methods.
One of the first examples of the application of BDA to clinical research. During the N1H1 influenza epidemic of 2008, the Google searches using the term “flu” or its symptoms proved very precise in predicting the increase in number of cases and the severity of the clinical condition. The image at top shows the number of searches, while the image at bottom shows the evolution of cases according to the Official Influenza Surveillance System in Spain (the time scales have been adjusted to ensure vertical correspondence between the two figures). This finding has subsequently been used as an epidemiological surveillance system, with great success.
Although to date it cannot be affirmed that BDA and ML have displaced the traditional clinical research methods, it is true that they allow the detection of trends or patterns in large bodies of data that may go unnoticed to the investigator, and which can guide the design of new studies adhered to the conventional methodology.30 Furthermore, they are the only alternative in those cases where it is logistically not possible to contemplate an orthodox clinical trial of sufficient statistical power to resolve a relevant question or issue.
Ethical and legal issuesIn a recent article appearing in Intensive Care Medicine, McLennan et al.31 offered an excellent analysis of the problems which the sharing of clinical data included in databases may pose. Since the coming into effect of the European Directive on General Data Protection Regulation (GDPR)32 in May 2018, the institutions must comply with legislation referred to the handling of personal data on patients in the European Union. The equivalent of this in the United States is known as Health Insurance Portability and Accountability Act (HIPAA), of 1996.33
The key issue is data anonymization: if the information is completely anonymous, there is no legal problem in sharing it from the right to privacy and security perspective. But when can we consider the data to be completely anonymous? Only when they do not lead to unequivocal identification of the patient upon being complemented with other data (e.g., even if we do not know the name of the patient or his or her history number, if we know the date of admission and age, we may identify the patient involved). We also must avoid unequivocal identification of the health staff appearing in the case history of the patient. In all other cases the data are considered to be pseudo-anonymized, and in this situation the GDPR directive would oblige us to request patient consent to use of the information.
Database anonymization is therefore crucial, and we must standardize a process that allows compliance with the European regulations while loosing as little information value as possible. Investigators must work jointly with the local Ethics Committees to harmonize the safeguarding of data privacy and security with the advances in clinical research which the BDA tools can offer us in future. Mechanisms must be developed to facilitate patient consent (or withdrawal of consent) to the use of the information generated during healthcare, conveniently harmonized for clinical research.
Another important issue is data property. In this case we are not dealing with privacy or security questions but with the intrinsic value of the information (even regarded as a commercial item). Does the patient (as the source of the information) or the clinician (as the subject entering the information in the system) have the right to request economical compensation or to prohibit the use of the data for clinical research purposes or any other use? Although the current reality is that the data are often in the hands of the companies that develop the medical software, and which seek to exploit such information for their own commercial interests,34 this issue is the subject of great controversy, and it would be desirable to establish legal norms seeking to resolve the problems we presently face in this field.
ConclusionsBig Data Analysis and Machine Learning tools offer a great opportunity for improving the strategic management of our Units, the handling of concrete clinical cases, and clinical research. However, in order to take advantage of this new methodology, we need to evolve and incorporate new human resources (specialized staff with clinical knowledge and training in AI) and technological assets. Furthermore, we must be able to harmonize the requirements referred to patient data privacy and security with the possibility of using large clinical databases efficiently.
We are convinced that an international and multidisciplinary team working collectively to draw knowledge from large bodies of clinical data, as explained in the course of this article, may bring a revolution to the modern practice of critical patient care.
Financial supportThe authors have received no financial support for the preparation of this article.
Conflicts of interestThe authors declare that they have no conflicts of interest in relation to the contents of this article.
Please cite this article as: Núñez Reiz A, Armengol de la Hoz MA, Sánchez García M. Big Data Analysis y Machine Learning en medicina intensiva. Med Intensiva. 2019;43:416–426.