The inflammatory response depends on several factors, including pathogenicity and duration of the stimulus, and also on the balance between the inflammatory and antiinflammatory response. Several studies have presented evidence of the importance of genetic factors in severe infections. The innate immune response prevents the invasion and spread of pathogens during the first few hours after infection. Each of the different processes involved in innate immunity may be affected by genetic polymorphisms, which can result in susceptibility or resistance to infection. The results obtained in the different studies do not irrefutably prove the role or function of a gene in the pathogenesis of respiratory infections. However, they can generate new hypotheses, suggest new candidate genes based on their role in the inflammatory response, and constitute a first step in understanding the underlying genetic factors.
La respuesta inflamatoria del huésped viene determinada por la virulencia del microorganismo, la duración del estímulo y el equilibrio entre la respuesta inflamatoria y la antiinflamatoria. Diversos estudios han mostrado la importancia de la genética en las infecciones graves. La respuesta inmune innata es el mecanismo que impide la invasión y propagación de microorganismos durante las primeras horas tras la infección. Cada uno de los procesos implicados en la respuesta innata puede alterarse por polimorfismos de los genes implicados, pudiendo esto resultar en una mayor susceptibilidad o resistencia a la infección. Los resultados obtenidos en los diferentes estudios genéticos no prueban de forma irrefutable el papel o la función de un gen en la patogénesis de la infección respiratoria. Sin embargo, permiten generar nuevas hipótesis, indican nuevos genes candidatos en base a su papel en la respuesta inflamatoria y proporcionan el primer paso en la comprensión de los factores genéticos subyacentes.
The genetic variability among individuals could explain why community-acquired pneumonia (CAP) can manifest as a serious illness in some people and as a relatively mild disorder in others. Sorensen et al.1 published a study on the causes of premature death in 1000 families with children adopted at an early age, and found that if the biological parents of these children had died as a result of infection before 50 years of age, the offspring had a relative risk of death due to infection of 5.81. In contrast, death of the adopting parents because of infection did not imply an increased risk of death due to infection in the adopted child. Thus, the susceptibility and response to infection appear to have a strong genetic influence.
In some diseases, the mutation of a single gene is necessary and sufficient to produce the clinical phenotype and cause the illness. These rare monogenic diseases are often associated to recurrent bacterial infections, and are detected in childhood.
However, most phenotypical traits of common diseases are determined by many genes that collaborate in different loci and lack the simple (or Mendelian) hereditary pattern that characterizes monogenic disorders. These complex or polygenic diseases are the result of the combination of different genetic and environmental factors, and include conditions as frequent as diabetes, hypertension, arteriosclerosis and susceptibility to infection. Such disorders exhibit a non-Mendelian hereditary pattern.
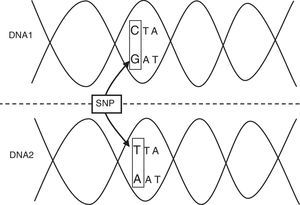
The incidence of infectious diseases indicates that the genetic variants associated to these processes must be relatively frequent (a polymorphism), rather than rare mutations. A genetic polymorphism is a region of the human genome (HG) that varies among different individuals in a given population. This allelic variant must affect a significant portion of the normal population (generally over 1%), and may involve the substitution of a single nucleotide (single nucleotide polymorphism, SNP) or affect the number of nucleotide simple sequence repeats (microsatellites), which account for over 50% of the HG.2,3 The so-called single nucleotide polymorphisms (SNPs) are the most important and frequent form of variation in the HG (Table 1; Fig. 1). Those polymorphisms that modify the sequence of amino acids encoded for by a gene or those bases located in its regulator region are the forms with the greatest probability of producing functional repercussions.2,3
Common genetic definitions.
| Mendelian inheritance | Transmission of hereditary characteristics according to the laws of Mendel. Hereditary pattern characteristic of monogenic diseases. |
| Single nucleotide polymorphism | Variation of the DNA sequence. A single nucleotide (A, T, C or G) in the genome differs among the members of a biological species or pairs of chromosomes. |
| Haplotype | Combination of allelic variants that tend to be inherited together. |
| Microsatellites | Repetitive short DNA sequences (simple sequence repeats). |
| Allele | One or more alternative forms of a DNA sequence. |
| Gene | A DNA segment involved in the production of a protein or RNA molecule. |
| Polymorphism | Difference in DNA sequence among individuals. |
| Exon | Any segment of a gene that is represented in the mRNA. |
| Intron | A non-encoding segment of the gene that is removed from the mature mRNA. |
| Heterozygote | Two different alleles in a given locus. |
| Homozygote | Two identical alleles in a given locus. |
| Linkage analysis | The identification of chromosomal regions associated to disease susceptibility, based on linkage studies in affected families. |
| Linkage disequilibrium | Two alleles in different loci that appear together in an individual more often than would be expected on a random (chance) basis. |
The innate immune system represents the first line of defense against the invasion and spread of pathogens in the first hours following infection. As a first step, the host must recognize the invading pathogen and induce its elimination through either complement-mediated lysis or phagocytosis. In turn, an inflammatory response must be triggered, and finally an antiinflammatory response is required in order to restore the homeostatic balance. Each of these processes can be affected by polymorphisms of the implicated genes, and which can produce susceptibility or resistance to infection. It also must be taken into account that although different genes within one same chromosome can determine susceptibility to infection on an independent basis, it is also possible that the observed association between a gene and a given disease only reflects what is happening in a neighboring gene. There is evidence that many alleles undergo en bloc segregation to form haplotypes. In this respect, while a SNP represents a variant of a single nucleotide, a haplotype consists of a considerably longer nucleotide sequence comprising several genes.2–4
Polymorphisms involved in antigen recognitionInnate immunity has developed a system for the recognition of common and constant molecular patterns on the surface of microorganisms, known as pathogen associated molecular patterns (PAMPs), through mediation of the so-called pattern recognition receptors (PRRs).
The main PAMPs that act as targets for activation of the innate immune system include physiological products of microbes such as lipopolysaccharides (LPS) (gramnegative species), lipoteichoic acid and peptidoglycan (grampositive species), zymosan (yeasts), DNA sequences with non-methylated CpG domains, and mannose or double-strand RNA (viruses). On the other hand, different types of proteins are able to recognize PAMPs. These PRRs include proteins of the complement system, such as mannose-binding lectin (MBL), and membrane receptors such as Toll-like receptors (TLRs) and CD14–expressed fundamentally at the surface of the cells that first come into contact with the pathogen during infection (cells of the epithelial surface), and by antigen-presenting cells (APCs) (monocytes/macrophages and dendritic cells).
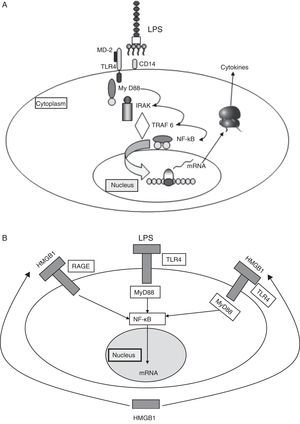
Binding of the PAMPs to PRRs triggers an intracellular signaling cascade that results in a series of antimicrobial processes and defensive functions.5 In the case of the TLRs, such signaling takes place through adaptor proteins. One of the most widely studied example is the myeloid differentiation factor 88, which binds intracellularly to a kinase associated to the receptor of interleukin-1 (IL-1). This in turn induces the activation of factor 6 associated to the receptor of tumor necrosis factor (TNF), causing the nuclear transcription factor NF-κB to translocate to the cell nucleus, where it activates the promoters of genes that encode for a broad range of pro- and antiinflammatory mediators (Fig. 2A).
(A) Signaling through TLR-4. TLR-4 recognizes LPS, an important component of the cell wall of gramnegative bacteria, and activates the innate immune system. The recognition of LPS requires CD14 in addition to TLR-4. The capacity of response of the TLR-4 and CD14 complex to LPS is reinforced by MD2. (B) Damage associated molecular patterns (DAMPs) (see text).
HMGB1: high-mobility group box 1; LPS: lipopolysaccharides; TLR: Toll-like receptor.
Although the PAMPs start and perpetuate the inflammatory response, there are other damage associated molecular patterns (DAMPs) which are released by the damaged cells and act as endogenous signals that induce and intensify the inflammatory response. These molecules constitute an important inflammatory mechanism, with an intensity of effect that depends on the type of damaged tissue and cell. The main DAMPs include different intracellular proteins, such as heat shock proteins (HSPs), high-mobility group box 1, and proteins derived from the extracellular matrix and which are generated as a result of tissue damage (Fig. 2B).
Lipopolysaccharide-binding proteinsLipopolysaccharides (LPS) are structural components of the bacterial wall, fundamentally of gramnegative species, and are considered to be key biological elements in the development of infectious and inflammatory processes. The binding of LPS to specific proteins such as LPS-binding proteins (LBPs), or permeability increasing protein, is implicated in phagocyte activation or neutralization.
Two groups of investigators6,7 have examined different LBP and permeability increasing protein polymorphisms, with conflicting results. In effect, while one group has found a polymorphism of the LBP gene (Cys98Gly) to be associated with an increased risk of sepsis in males and a poorer prognosis,6 the other group was unable to reproduce these results.7 Recently, Flores et al.8 have identified an LBP gene haplotype associated with susceptibility to serious infection.
CD14CD14 is a membrane protein expressed by macrophages and monocytes and, in lesser proportion, by neutrophils. In addition to binding to lipid A of LPS, CD14 can also bind to the peptidoglycan of Staphylococcus aureus, to lipoarabinomannan of mycobacteria, and to other components of the wall of streptococci. A SNP has been described, located in position−159 of the promoter region of the gene (chromosome 5), affecting a cytosine–thymine transition, and which in individuals homozygous for the T allele gives rise to increased circulating soluble CD14 levels and an increased production of interferon-gamma (IFN-γ). The CD14−159 TT genotype was seen to appear more often in a population of septic shock patients compared with a control group of healthy individuals, and was moreover associated to increased mortality.9 Other authors have been unable to corroborate these findings, however.10
Toll-like receptorsThere are at least 11 TLRs with different affinities for different microbial antigens. TLR-4 appears to be essential in recognizing the endotoxin, while TLR-2 is more important in recognizing peptidoglycan of grampositive bacteria. The activation of TLRs implies an increased expression of molecules of the major histocompatibility complex (MHC), an increased expression of co-stimulator molecules, and an increase in the expression of genes dependent upon NFκB, such as IL-1, IL-6, IL-12 and TNF-α. Several SNPs of TLR-4 and TLR-2 (chromosome 9) have been identified that appear to increase the risk of serious bacterial infections–though the existing data are conflicting in this respect.10–12 Another study has found variability in the TLR-5 gene to affect the capacity to recognize flagellin, and carriers of haplotypes TLR-5 1174* 2-1175* 1 and TLR-5 1174* 1-1175* 2 are at an increased risk of infection due to Legionella pneumophila13 (Table 2).
Association between single nucleotide polymorphism in genes related to immunity and community-acquired pneumonia.
| Single nucleotide polymorphism | Association to community-acquired pneumonia | References |
| Mannose-binding lectin | ||
| MBL2 Gly54Asp (G/A)-rs1800450 (allele B) | Allelic variant associated to risk of invasive pneumococcal disease | Moens et al.14 |
| MBL2 Gly57Glu (G/A)-rs1800451 (allele C) | ||
| MBL2 Arg52Cys (C/T)-rs5030737 (allele D) | Allelic variants associated to increased severity and poorer prognosis | |
| Alleles B, C and D and −221 G/C-rs7096206 (alleles X/Y) | No association to CAPNo association to CAP-P | Garcia-Laorden et al.15 |
| Fcγ II receptors | ||
| FCGR2 Arg131His (C/T)-rs1801274 | Homozygotic allele 1 is a risk factor for pneumococcal bacteremia | Yuan et al.19 |
| Homozygotic allele 1 is associated to increased severity of CAP | Endeman et al.18 | |
| Homozygosis for variant H predisposes to pneumococcal bacteremia | Solé-Violán et al.20 | |
| Toll-like receptors | ||
| TLR2-16934 (T/A)-rs4696480 | Homozygotic allele 2 is associated to increased risk of pneumonia | Sutherland et al.9 |
| TLR2 Arg677Trp (C/T)-rs5743706 | Carriers of allele 2 are at increased risk of sepsis due to gramnegative bacteria | Lorenz et al.12 |
| TLR2 Arg753Gln (G/A)-rs5743708 | No association to CAP | Yuan et al.19 |
| TLR4 Asp299Gly (A/G)-rs4986790TLR5 Arg392 (C/T)-TLR5 Asn592Ser (A/G) | Carriers of allele 2 and haplotypes TLR5 1174* 2-1175* 1 and TLR-5 1174* 1-1175* 2 are at increased risk of infection due to Legionella pneumophila | Hawn et al.13 |
| Tumor necrosis factor | ||
| TNFA-308 (G/A)-rs1800629 | No association to CAP | Solé-Violán et al.25 |
| TNFA-238 (G/A)-rs361525TNFA-308 (G/A)-rs1800629 | Carriers of allele 2 are at increased risk of poor outcomes | Kinder et al.21 |
| TNFA-238 (G/A)-rs361525TNFRSF1B+676 (G/T)-rs1061622 | Haplotype TNF308*2-238*2 is associated to increased mortality | Henckaerts et al.22 |
| Met196Arg | Heterogeneity protects against poor outcome in CAP | Solé-Violán et al.25 |
| Interleukin-6 | ||
| IL6-174 (C/G)-rs1800795 | Homozygosis for allele G protects against poor outcome in pneumococcal CAP | Martín-Loeches et al.29 |
| Surfactant proteins | ||
| SFTPA1 (aa19T/C)-rs1059047 | Haplotypes SFTPA2 1A10 and SFTPA1-SFTPA2-6A3-1A predispose to CAP | García-Laorden et al.36 |
| SFTPA1 (aa50 G/C)-rs1136450 | Haplotypes A10 and 6A-1A are associated to poor prognosis | |
| SFTPA1 (aa219C/T)-rs 4253527 | ||
| SFTPA2 (aa9 A/C)-rs1059046 | ||
| SFTPA2 (aa91 G/C)-rs17886395 | ||
| SFTPA2 (aa223C/A)-rs4253527 | ||
| SFTPD (aa11T/C)-rs721917 | ||
| Heat shock proteins | ||
| HSP70-2+1267 AA-rs1061581 | Genotype HSP70-2 1267 predisposes to septic shock in patients with CAP | Waterer et al.34 |
Mannose-binding lectin (MBL) is a molecule of the innate immune system that is able to activate a complement upon binding to different microbial surface sugar molecules. In addition, MBL can act directly as an opsonin and bind to specific receptors expressed on the surface of different types of cells. This protein is encoded for by a single gene (mbl2) located in chromosome 10, and there are three structural allelic variants, along with others in the promoter region of the gene that significantly reduce its plasma levels.14 We have studied different MBL polymorphisms in patients with community-acquired pneumonia (CAP), and have found MBL-deficient allelic variants to be associated with an increased risk of developing severe forms of the disease and a poorer patient prognosis.15 However, these polymorphisms did not confer increased susceptibility to CAP15 or to pneumococcal pneumonia.16 The existence of linkage disequilibrium between the MBL genes and the surfactant proteins indicates that the genetic variability of the latter could be responsible for the susceptibility to pneumococcal infection observed in other studies (Table 2).17
Receptors for the Fc portion of immunoglobulin GLeukocyte receptors for the Fc portion of immunoglobulin G (FcγR) are essential for the defense against certain microorganisms. The binding of IgG to these receptors activates the complement system, facilitates phagocytosis and activates cellular cytotoxicity. The substitution of histidine with arginine in position 131 of the amino acid sequence of the gene encoding for the FcγRIIa receptors produces a decrease in their affinity for IgG2, and alters the host defenses against encapsulated microorganisms.18,19
In patients with CAP, Solé-Violán et al.20 have found homozygosity for variant H131 that is predisposed to bacteremic pneumonia and a poorer outcome (Table 2).
Polymorphisms implicated in the inflammatory responseBinding of PAMPs to PRRs results in the activation of NF-κB, an intracellular protein that is translocated to the nucleus, inducing the transcription of proinflammatory cytokines. The inflammatory reaction is a key element in the host defense mechanisms.
Tumor necrosis factorTumor necrosis factor-alpha (TNF-α) plays a key role in the acute inflammatory response following an infectious stimulus, and its levels of expression are therefore of potential relevance to the clinical course. A number of SNPs have been described in the locus corresponding to TNF (chromosome 6).21,22 The most extensively studied of these is TNF-α-308–its allele A (transition of guanine to adenine) being associated to significantly increased production of the protein. This polymorphism has been associated to a poorer prognosis in septic shock patients,23 though here again other studies have obtained opposite results (Table 2).24,25 This can be explained by taking into account that this polymorphism is in linkage disequilibrium with other SNPs of the promoter region and other polymorphisms of neighboring genes–many of which also play an important role in the inflammatory response. This is the case of allele A of lymphotoxin-alpha (LTA + 250 A), which is almost always associated to an allele G in position TNF-α-308. The LTA + 250 AA (transition of guanine to adenine in the first intron) genotype has been associated to increased TNF levels and to an increased risk of septic shock in patients admitted to hospital due to CAP. In contrast, respiratory failure in the absence of septic shock was closely correlated to the LTA + 250 GG genotype.26 The proximity of the TNF and LTA genes to other immunologically important genes in the adjacent region of chromosome 6, such as the loci for HLA, complement and HSP, may further complicate the evaluation of association studies.
Interleukin-6Interleukin-6 (IL-6) is a prognostic marker in sepsis, though the underlying causal relationship is not clear. The GG genotype of polymorphism IL-6-174 has been associated to increased survival in patients with severe sepsis.27 A haplotype conferring increased mortality risk and organ dysfunction in patients admitted to the Intensive Care Unit (ICU) has also been identified.28 However, other authors have not found this polymorphism to be associated to differences in prognosis (Table 2).29
We have explored the clinical repercussions of the genetic variants of IL-6 in 1227 patients with CAP. In the subgroup of individuals with pneumococcal CAP, genotype IL-6 −174 GG was associated to lesser severity and improved prognosis.29 IL-6 plays an important role in the host immune defense against pneumococcal pneumonia, and is the main inducer of the synthesis of acute phase reactants by the liver cells–particularly complement factor C3 and C-reactive protein. The latter in turn binds to the polysaccharides of the cell wall of Streptococcus pneumoniae, facilitating its recognition by the host immune cells.30–32
Polymorphisms implicated in the antiinflammatory responseThe evolution of the inflammatory response depends on a number of factors, including the virulence and duration of the stimulus, and also the balance between the inflammatory and antiinflammatory responses. The antiinflammatory cytokines are responsible for the down-regulation of cellular and humoral immunity, inducing a period of relative immune suppression referred to as compensatory antiinflammatory response syndrome. The genetic polymorphisms responsible for an intense or prolonged compensatory antiinflammatory response syndrome may have the same dramatic consequences as an uncontrolled inflammatory response.
Interleukin-1 receptor antagonistThe IL-1 receptor antagonist (IL-1RN) represents the physiological antiinflammatory opponent to IL-1, and thus protects against the adverse consequences of an excessive inflammatory response. There is a polymorphic region in intron 2 of the IL-1RN gene that contains a variable number of tandem repetitions. The alleles are called A1, A2, A3, A4 and A5, according to the range of frequencies they present in the healthy population. Allele IL-1RNA2 is associated to an increased production of IL-1RN following stimulation with LPS, and is more common in patients with severe sepsis–though it does not appear to imply a poorer prognosis.33 Patients who are homozygous for LTA+250 A and IL-1RNA2 appear to be at an increased risk of death due to sepsis, though these data have not been confirmed by a recent study.18
Pathways activated by inflammationDamage associated molecular patternsA number of molecules, including high-mobility group box 1 and the HSPs, constitute “alarm signals” that stimulate the inflammatory response. In this respect, SP70-2 is an important immune modulating protein induced in response to inflammatory stimuli. In a study of 343 patients admitted due to CAP, Waterer et al.34 found genotype HSP70-2 1267 AA to be associated to an increased risk of septic shock–carriers of haplotype HSP70-2+1267A/LTA+250A being the individuals at highest risk.34
Surfactant proteinsSurfactant proteins (SPs) are colectins secreted by type II pneumocytes. During acute pulmonary infections, these molecules can destroy, opsonize and/or stimulate the phagocytosis of microorganisms and modulate pulmonary inflammation. Different polymorphisms of the genes encoding for SP-A, B, C and D are known. Genotype SP-B+1580CC involves a thymine/cytosine variation in position 1580 in exon 4, resulting in a change from threonine to isoleucine in the amino acid chain of the protein, and gives rise to a functional alteration of SP-B. An association has recently been demonstrated between this SNP and the risk of septic shock and respiratory failure in patients with CAP (Table 2).35
García-Laorden et al.36 evaluated the association between the genetic variability of SPs and the susceptibility to and prognosis of CAP. Allele SFTPD aa11-C was significantly associated to lesser serum levels of SP-D. Furthermore, several SFTPA1, SFTPA2 and SFTPD haplotypes were associated to increased susceptibility to CAP, and to a poorer patient outcome.
Coagulation proteinsInflammation and coagulation are intimately related. Tissue factor, produced by neutrophil adhesion and cell damage, activates the coagulation cascade and stimulates the inflammatory response. Severe pneumonia is characterized by an important inflammatory response that stimulates procoagulant activity and reduces fibrinolysis. Plasminogen activator inhibitor type 1 (PAI-1) is the main inhibitor of the fibrinolytic system.
A polymorphism has been identified in the gene encoding for PAI-1–high plasma levels of the latter being associated to hypercoagulability states. Pneumonia patients carrying allele 4G are at an increased risk of suffering septic shock and multiorgan dysfunction.37 Another study found the PAI-1 genotypes associated with increased circulating levels of the protein to be correlated to increased susceptibility to CAP.38
The future of genetic studies in infectionsThe growing availability of information on SNPs in the HG, and the great diversity of clinical phenotypes to which they may be related, has stimulated the conduction of many studies on the association between polymorphisms of candidate genes and different phenotypes of common illnesses. However, very few of the published associations have been unequivocally reproduced by other investigators. The limited sample sizes and inadequate matching of patients with the controls may have contributed to this situation.
Methodologically solid genetic association studies involving a case-control design, pangenomic studies,39 and the analysis of the differential expression of genes will allow us to identify new candidate genes on the basis of their role in the inflammatory response, and will help to clarify the molecular events required for a pathogen to be able to invade the host, and the host mechanisms involved in eliminating it. The results of such research undoubtedly will cause a revolution in the development of vaccines and antimicrobials.40 Lastly, the study of specific pharmacogenetic profiles will allow us to identify patients with different possibilities to respond to certain drugs, and to better adjust the prescribed antimicrobial treatments.
Conflicts of interestThe authors declare that they have no conflicts of interest.
Please cite this article as: Ferrer Agüero JM, Millán S, Rodríguez de Castro F, Martín-Loeches I, Solé Violán J. Neumonía adquirida en la comunidad: variantes génicas implicadas en la inflamación sistémica. Med Intensiva. 2014;38:315–323.