Streptococcus pneumoniae and Neisseria meningitidis are major causes of severe invasive bacterial infections in some individuals. Apparently the genetic is a major susceptibility determinant to these infectious diseases. We study if the functional polymorphisms within genes of the innate immune system (TLR2–TLR4 and CD14) are related to the predisposition to severe invasive infections caused by S. pneumoniae and N. meningitidis.
Material and methodsProspective descriptive study. Sixty-six Caucasian healthy children and 173 consecutive Caucasian children with invasive bacterial infections by N. meningitidis (n=59) and S. pneumoniae (n=114) were enrolled between January 1, 2008 and December 31, 2010. All blood samples were genotyped with description of the coding polymorphisms in p.R753Q of TLR2 gene and p.D299G of TLR4 gene as well as the promotor polymorphism c.-159C>T of the CD14 gene.
ResultsCompared to the controls the p.753Q allele of TLR2 and the allele c.-159T of CD14 were more frequent in patients with S. pneumoniae (p<0.0001 and p=0.0167) and meningococcal infections (p=0.0003 and p=0.0276 respectively).
ConclusionsGenetical variations in the innate immune system by polymorphisms in the TLR2 and CD14, could be related with an increases susceptibility to severe invasive infections by S. pneumoniae and N. meningitidis.
Streptococcus pneumoniae y Neisseria meningitides son causantes de infección bacteriana grave en algunos individuos. Cierta susceptibilidad genética puede ser determinante para este hecho. Nuestro objetivo es determinar si el polimorfismo de genes relacionados con el sistema inmune innato (Toll like receptor 2 y 4 junto con CD14) se relaciona con la predisposición a sufrir infecciones graves por los citados patógenos.
Material y métodosEstudio prospectivo observacional (desde el 1 de enero de 2008 hasta el 31 de diciembre de 2010). Se incluye a 66 niños sanos y 173 niños con infección bacteriana grave (59 por Neisseria meningitidis y 114 por Streptococcus pneumoniae). Todas las muestras fueron genotipadas para los polimorfismos p.R753Q de TLR2, p.D299G de TLR4 y c.–159C>T del CD14.
ResultadosComparados con los controles, los polimorfismos p.753Q de TLR2 y c.–159C>T de CD14 fueron más frecuentes en pacientes con infección neumocócica (p<0,0001 y p=0,0167) y meningocócica (p=0,0003 y p=0,0276).
ConclusionesLas variaciones genéticas en el sistema inmune innato mediante polimorfismos en TLR2 y CD14 podrían estar relacionadas con la susceptibilidad a las infecciones graves por Streptococcus pneumoniae y Neisseria meningitides.
It is known that Streptococcus pneumoniae and Neisseria menigitidis are causes of severe invasive bacterial infections in some individuals, producing high morbidity and mortality, leading to mild or banal infections in others. The asymptomatic nasopharyngeal colonization by these bacterias is common and is related with an invasive disease in only a short number of cases.1,2 It has been described that the rate of carriers of Neisseria meningitidis rise to 80% in severely crowded conditions.3,4 On one hand, the instauration of S. pneumoniae conjugate vaccine did not decrease as expected the rates of nasopharyngeal carriers for S. pneumoniae.5,6
There are multiple underlying immune defects that may predispose to invasive infections as inmunosuppression (primary or secondary), asplenia or immune factors deficiency (properdin, components of complement or MyD887,8). Individuals with these conditions have an increased risk but they are only a small proportion of all cases.
In healthy patients the innate immune system looks crucial for the early containment of microbial infections by triggering inflammation and coordinating the acquired immune response. The family of Toll-like receptors (TLRs) is a central component of this system and its description has permitted a better understanding of the molecular mechanisms concerning to antimicrobial and inflammatory responses. The TLRs seem to play a key role in signaling molecules of pathogens and endogenous proteins related to immune activation given their ability to recognize evolutionary conserved pathogen-associated molecular patterns of microbial origin.9
Several TLRs have been identified in humans, and each one recognizes different structures, also each bacterium could activate different sets of TLRs. The TLR2 is activated by bacterial lipoproteins,10 peptidoglycan and lipoteichoic acid of the cell wall. It is well known that the ability of S. pneumoniae to activate TLR2 through this pathway.9,11,12 It has been also described that S. pneumoniae pneumolysin could also stimulate cells through TLR4, but this observation has not been confirmed by all studies.13 Moreover, besides TLR2, and maybe TLR4, additional TLRs are probably involved in recognition of this Gram-positive bacteria, among them TLR9.9,14
Lipopolysacharide (LPS) is a major component of the outer cell wall of Gram-negative bacteria such as N. meningitidis. The immune cell recognition of LPS involves an LPS receptor complex; of which CD14 and TLR4 are important components15 and its activation could depend from the bacterial dosage. At lower bacterial concentrations, LPS activation of macrophages is TLR4/CD14 dependent. This activation can be blocked by specific antibodies to CD14 and TLR4.16 Higher bacterial concentrations activate the complement and non-LPS components initiate TLR2 and TLR4 activation independent of CD14.17 Finally the porin, an outer membrane protein of N. meningitidis stimulates immune response also through TLR2.18,19
Genetic variation of immune response genes is associated with susceptibility to and severity of infectious diseases.20–22 Single nucleotide polymorphisms could be in the origin of this risk explaining the interindividual susceptibility to invasive bacterial infections. In the present study we analyze the presence of a set of polymorphisms (p.R753Q of TLR2, p.D299G of the TLR4 gene and c.-159C→T located in the CD14 promoter) in 157 children with severe invasive bacterial infection by S. pneumoniae and N. meningitidis. We hypothesize that severe invasive bacterial infections by these microorganism affect the susceptible healthy individuals. Their susceptibility could be determined by genetic factors based on the relationship between its variants and the susceptibility to these infections.
Materials, patients and methodsControls and cases characteristicsThe study was approved by the ethics committee of the Hospital Infantil Universitario Niño Jesús of Madrid (Spain) according to local laws and regulations. The patients were recruited from January 1 (2008) to December 31 (2010) after signing the consent by the legal tutors or caregivers of each patient.
Sixty-six Caucasian healthy children were enrolled as controls: the samples were collected from blood samples taken in the hospital blood-extractions department because of non-infectious or inflammatory causes.
The cases were collected from the Pediatric Intensive Care Unit (PICU). One hundred and seventy three consecutive previously healthy Caucasian children were enrolled because an invasive bacterial infection by N. meningitidis (n=59) or S. pneumoniae (n=114).
The criteria applied for invasive meningococcal infection were:
- •
Isolation of N. meningitidis in blood or cerebrospinal fluid.
- •
Presence of Gram-negative diplococcus in cerebrospinal fluid.
- •
Severe sepsis and extensive purpura without identification of causing agent.3,4,23
The criteria for invasive S. pneumoniae infection were:
- •
Isolation of Gram-positive diplococcus in culture or positive protein chain reaction (PCR) for S. pneumoniae in a sterile body fluid (blood, cerebrospinal fluid, pleural fluid or peritoneal fluid) or detection of S. pneumoniae antigen by immunochromatography in the same fluids (Binnax Now®).
Blood samples from cases were obtained at hospital blood extractions department. For the cases the blood samples were obtained at PICU admission and submitted for genotyping to the laboratory of genetics. The DNA was extracted and genotyped by staff blinded to clinical data.
All polymorphisms were genotyped by restriction analysis after PCR amplification. PCR of 35 cycles was performed on a thermal cycler GeneAmp9700 (Perkin-Elmer Cetus, Norwalk, CT, USA). Primers were synthesized (VWR International Eurolab, Bcn, Spain). The PCR reaction consisted of 50ng DNA, 10pm of each primer, 10μl PCR master mix (Promega®, Madison, WI, USA) and water up to 20μl.
- 1
TLR2 p.R753Q (rs5743708) PCR primers were 5′-GAAGAGAACAATGATGCTGCCATTC-3′ and R: 5′-CTAGGACTTTATCGCAGCTCTC-3′. The cycle program consisted of 94°C for 30s, 49°C for 30s, and 72°C for 30s. SsiI (Fermentas, Burlington, Canada) digestion resulted in two fragments of 115bp and 58bp (R allele) or 173bp (Q allele).
- 2
TLR4 p.D299G (rs4986790) PCR primers were 5′-ACTTAGACTACTACCTCGGTG-3′ and R:5′-GATTTGAGTTTCAATGTGGGAAAC-3′. The cycle program consisted of 94°C for 30s, 53°C for 30s, and 72°C for 30s. HphI (Fermentas, Burlington, Canada) digestion resulted in two fragments of 168bp and 15bp (G allele) or 183bp (D allele).
- 3
CD14 c.-159C>T (rs2569190) PCR primers were: 5′-TCACCTCCCCACCTCTCTT-3′ and R: 5′-CCTGCAGAATCCTTCCTGTT-3′. The cycle program consisted of 94°C for 30s, 59°C for 30s, and 72°C for 30s. HaeIII (Roche® Mannheim Germany) digestion resulted in two fragments of 85bp and 22bp (C allele) or 107bp (T allele).
Statistical analysis was performed in a Hewlett–Packard computer with the statistical program SPSS® 19.0 (IBM®). A descriptive analysis was done with the epidemiological and clinical variables; the values are shown with mean and range. Genotype and allele frequencies between groups were compared using the chi-square test. Alternatively Fisher's exact test was used when frequencies were <5. The value reported is the Yates chi-square, corrected for continuity when applicable. Findings of p<0.05 were considered statistically significant.
ResultsControls and cases characteristicsThe control group has a mean age of 50 months (range 3 months–14 years); 33 were males and 33 females. The mean age of the patients was 41 months (range 3 days–17 years); 92 of them were males and 81 females.
The distribution of severe invasive bacterial infection by N. meningitidis was: meningitis n=8, sepsis n=41 and meningitis with sepsis n=10.
The distribution for S. pneumoniae infections was: meningitis n=12, Sepsis n=102.
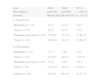
GenotypingGenotypic distribution of the polymorphisms studied in patients and controls are shown in Table 1.
Genotypic distribution of the studied polymorphsims in patients and controls. Patients have been sudivided depending on the phenotype.
| Gene | TLR2 | TLR4 | CD 14 |
| Polymorphism | p.R753Q | p.D299G | c.-159C>T |
| Genotype | RR-RQ-QQ | DD-DG-GG | CC-CT-CT |
| S. Pneumoniae | |||
| Meningitis (n=12) | 4-7-1 | 9-3-0 | 1-6-5 |
| Sepsis (n=10) | 5-3-2 | 9-1-0 | 3-4-3 |
| Pneumonia and sepsis (n=92) | 37-46-9 | 76-13-3 | 13-48-31 |
| Total (n=114) | 46-56-12 | 94-17-3 | 17-58-39 |
| N. Meningitidis | |||
| Meningitis (n=8) | 5-1-2 | 7-0-1 | 3-3-2 |
| Sepsis (n=41) | 15-19-7 | 29-9-3 | 5-25-10 |
| Meningitis and sepsis (n=10) | 4-4-2 | 9-0-1 | 1-8-2 |
| Total (n=59) | 24-24-11 | 45-9-5 | 9-36-14 |
| Controls (n=66) | 49-15-2 | 60-4-2 | 21-31-14 |
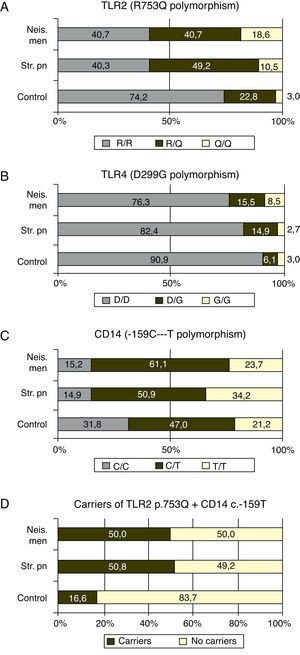
The p.753Q allele of the p.R753Q polymorphism of the TLR2 gene was clearly overrepresented in patients compare to controls. Fifty nine percent of the patients with meningococcal infection (p=0.0003) and 59.3% of those with S. pneumoniae infection, (p<0.0001) carried at least one copy of this allele (see Fig. 1 and Table 2).
p values of the comparison of genotypic frequencies of the polymorphisms studied between patients with severe invasive infections caused by Streptococcus pneumoniae and Neisseria meningitidis and controls under the hypothesis of codominance, dominance or recessivity of the wild-type allele.
| TLR2 p.R753Q | TLR4 p.D299G | CD14 c.-159C>T | |
| N. meninigitidis vs. controls | |||
| Codominance | p: 0.0003 | n.s | n.s |
| Dominance of the WT allele | n.a. | n.a. | n.s |
| Recessivity of the WT allele | p: 0.0003 | p: 0.0472 | p: 0.0276 |
| S. pneumoniae vs controls | |||
| Codominance | p: <0.0001 | n.s | p: 0.0167 |
| Dominance of the WT allele | n.s | n.s | n.s |
| Recessivity of the WT allele | p: <0.0001 | n.s | p: 0.0128 |
| N. meninigitidis. vs. S. pneumoniae | |||
| Codominance | n.s | n.s | n.s |
| Dominance of the WT allele | n.s | n.s | n.s |
| Recessivity of the WT allele | n.s | n.s | n.s |
The frequency of carriers of the p.299G allele was higher (p=0.0472) in the case of meningococcal infection (23.7%) compared to controls (9.1%). The allelic frequency of the p.299G allele was also higher (p=0.0189) in this group of patients (16.1%) when compared to healthy children (6.1%) with (data not shown in tables). No differences were found between patients with S. pneumoniae infections and controls (see Fig. 1 and Table 2).
CD14The study of c.-159C>T polymorphism showed that the allele c.-159T was more frequent in both group of patients than the c.-159C allele which was the most frequent in controls. Nevertheless, the p value reached statistical significance only comparing the allelic frequencies in patients with meningococcal infections and controls (p=0.0084). The frequency of patients with one or two copies (carriers) of the allele c.-159T was higher in case of meningococcal (86.4%; p=0.0276) or S. pneumoniae (85.1%; p=0.0128) infections than in controls (68.2%; see Fig. 1 and Table 2).
TLR4 plus CD14Taking together the p.753Q of TLR2 and c.-159T of CD14 alleles, 50.8 ((30/59) and 50.0% (57/114) of the patients with meningococcal and S. pneumoniae infections carried at least one copy of both risk alleles (Fig. 1D) while this haplotype was found in 16.6% (11/66) controls (p<0.0001 in both cases)
Hardy–Weinberg equilibriumThe distribution of genotypes in the control population was in H–W equilibrium in all the studied polymorphisms.
DiscussionThe ability to sense pathogenic organisms and to respond adequately preserving the host biological identity is essential to survival. The innate immune system is the first defence line against invading pathogens. It plays a key role in acute host response.24 The pathogens are recognized by a set of receptors which recognize conserved pathogen associated molecular patterns (PAMPs). The potential effect of the genetic variability on the individual susceptibility to disease is not completely known. Related to this, in our study, there are two main original findings: (1) patients with N. meningitidis or S. pneumoniae infections showed a higher prevalence of p.753Q TLR2 variant and c.-159T allele of the CD14 promoter polymorphism; (2) the ratio of carriers for the p.299G allele was slightly higher in meningococcal disease.
TLR2TLR2 can recognize a wide spectrum of PAMPs including molecules shared by Gram-positive bacteria or specific molecules of Gram-negative bacteria (as the porins of meningococci or the LPS of Bordetella or Legionella species). Moreover, TLR2 is capable to recognize molecular patterns of viruses, parasites and mycobacteria.
Association between TLR2 polimorphisms and leprosy, tuberculosis and staphylococcal infection has been previously reported. Moreover TLR2-knockout mice are more susceptible to septicemia and/or meningitis caused by a wide spectrum of bacteria including S. pneumoniae.25,26 Also TLR2-deficient mice show an increased susceptibility to infections, suggesting that polymorphisms that affect TLR expression or function may impair host response to a given spectrum of pathogens.
TLR2 is probably the most important receptor for Gram-positive bacterial products. Moreover, TLR2 is capable to detect a wide variety of molecules from Gram-negative bacteria, between others the porin from meningococci. This ability to recognize such a variety of ligands probably arises from its potential to form TLR2 homodimers and heterodimers with TLR1 and TLR6.27
In our study, the ratio of patients with N. meningitidis or S. pneumoniae infection that carried the allele p.753Q were close to 60%, much more than the expected 25% based on the frequency found in the control group (see Fig. 1A). These findings suggest that the p.753Q allele affects the ability of TLR2 to respond to molecules of S. pneumoniae. This finding agrees to the observation that transfected cells with p.753Q TLR2 variant show impaired cellular activation in response to lipoproteins.
Our data suggest strongly that the p.753Q allele also affects the host response to N. meningitidis. This polymorphism is within the TIR domain of TLR2 which is critical to TLR signaling and dimerization with other TLR molecules24,28 and have been linked to decreased NF-κB activation and to increased risk of infection.29 The impaired dimerization of TLR2 could be linked to a decreased response to meningococcal porin PorB which requires TLR1 for signaling.30
TLR4The macrophages activation by meningococcal lipopolysaccharide (LPS) is TLR4 dependent. The p.D299G substitution is associated with functional changes as demonstrated by impaired airway responsiveness after LPS stimulation.30
In this study we found that the ratio of carriers for the p.299G allele was slightly higher in the group of patients with meningococcal disease, as well as the allelic frequency of this transition (p=0.0189, data not shown). Our study did not show association between S. pneumoniae disease and this TLR4 polymorphism.
While some previous studies have not found association between this polymorphism and meningococcal disease31–33 there is a previous study34 that reports a significantly higher incidence of Gram-negative infections in a cohort of patients with systemic inflammatory response syndrome bearing the p.299G allele of TLR4.
CD14The CD14 is the anchor protein of the TLR4 receptor complex and its soluble form binds to Gram-negative LPS and the antibodies to CD14 blocks meningococcal LPS activation of macrophages.35 Moreover, CD14 is a high affinity receptor for bacterial endotoxins, constituents of bacterial cell-wall. Several studies confirmed that CD14 interacts not only with LP from Gram-negative bacteria, but also with other microbial ligands as lipoteichoic acid and peptidoglycan from Gram-positive bacteria.16
The promoter polymorphisms c.-159C>T included in our study had been previously associated with shock and mortality rate in patients with sepsis; it causes a reduced circulating CD14 concentrations.36 In our study, 86.4% (51/8) and 85.1% (97/114) of the patients with meningococcal and S. pneumoniae infections respectively carried the c.-159T allele of the CD14 promoter polymorphism. In the control group, the rate of carriers reaches 68.2% (45/66) which is significantly lower.
The broad specificity of CD14 in ligand recognition suggests that a decreased CD14 response linked to the c.-159T allele of this promoter polymorphism could impair the recognition and binding of different microbial endotoxins (LPS to CD14). It could decrease the triggering of a signaling cascade-mediated by Toll-like receptors that promote the synthesis of multiple host-derived inflammatory mediators. Although the trend seems to be similar, the p values obtained are more significant for S. pneumoniae than meningococcal infections; this observation could be due to the shorter number of patients in the second group.
Finally, the effect of the “risk” alleles of TLR2 and CD14 is additive: only 16% of the controls bore at least one copy of both alleles, while this haplotype was found in 50.0 and 50.8% of the patients respectively with a p=0.0001.
This study has limitations; first the small numbers of controls and patients. Also the clinical evolution and cases outcome were not collected and analyzed; it should be done in future studies in order to consider the influence of these findings in the patients’ evolution. The statistical approaches to multiple hypothesis testing and lack of validation of findings in a second cohort owe to interpret the significance of the results with precaution.
ConclusionCorrelation between functional polymorphisms within the TLR2, TLR4 and CD14 and children susceptibility to severe invasive bacterial infections is not completely defined. In our study the c.-159T allele of the CD14 gene and, especially, the p.753Q allele of TLR4 could be related to an increased risk of developing severe infections by S. pneumoniae and N. meningitidis. Our data suggest a key role of the innate immunity system in the protection against these bacterias that should be confirmed in future studies.
Conflict of interestThis work has been supported by grant by the “Fondo de Investigaciones Sanitarias” from the Institute Carlos III.
AbbreviationsLipopolysacharide: LPS.
Pediatric Intensive Care Unit: PICU.
Protein chain reaction: PCR.
Toll-like receptors: TLRs.
Thanks to our patients and to all the professionals who work in the pediatric critical care unit.
This work has been supported by grant by the “Fondo de Investigaciones Sanitarias” from the Institute Carlos III.